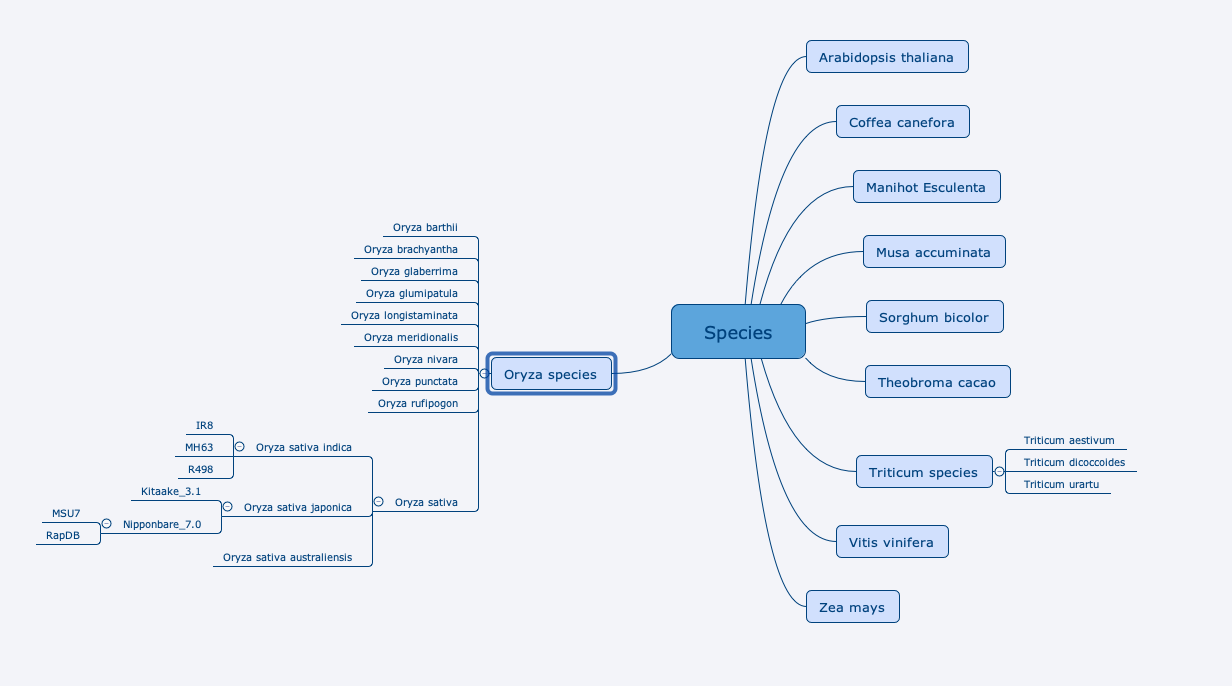
This page provides a summary on the species, data sources and URI patterns.
AgroLD includes data on the following species on:
Ontologies in AgroLD:
The OWL versions of the ontologies have been loaded to AgroLD. The original namespaces and URIs have been retained.
Data sources in AgroLD:
| Data source | Information | Website | Notes | Species | Ontologies used |
|---|---|---|---|---|---|
| Ensembl plants | Gene, Orthologs | plants.ensembl.org | This resource host several genome of plant species and gather their annotations | All | GO, SO, RO, SIO |
| Gramene | Gene, QTL, Pathways and ontology associations (PO, TO and EO) | www.gramene.org | In the future versions, ontology associations will be taken from the Planteome project (http://planteome.org/) | R,A, W, S, M | GO, PO, TO, EO |
| GOA | Gene Ontology associations | www.ebi.ac.uk/GOA | All | RO,GO,SIO | |
| UniprotKB | Protein Information | www.uniprot.org | All | GO,RO,SIO | |
| GreenPhylDB | Web resource for phylogenetic-based approach to predict homologous relationships. | www.greenphyl.org | R,A | GO,RO,SIO,ORTH | |
| Oryza Tag Line | Database consists of phenotypic data resulting from the evaluation of the Génoplante rice insertion line library. | oryzatagline.cirad.fr | R | PO, TO | |
| Rice Genome Hub | Genes, annotations, Ontology associations | rice-genome-hub.southgreen.fr | R | GO,SIO,FALDO,RO,SO | |
| TropGeneDB | Database for genomic, genetic and phenotypic information about tropical crops. | tropgenedb.cirad.fr | R | GO, TO, PO | |
| Oryzabase | Genes, ontology associations, publications | shigen.nig.ac.jp/rice/oryzabase | R | GO,PO,TO | |
| Interpro | Classification of protein families | www.ebi.ac.uk/interpro | All | GO | |
| RAPdb | The Rice annotation project | rapdb.dna.affrc.go.jp | R | GO,SIO,FALDO,RO,SO | |
| MSU RGAP | MSU Rice genome annotation project | rice.plantbiology.msu.edu | R | GO,SIO,FALDO,RO,SO | |
| RiceNetDB | Gene networks database | bis.zju.edu.cn | R | GO | |
| StringDB | Protein-Protein interactions network database | string-db.org | R,A | GO |
Datasets:
Datasets releases are available through Zenodo under CC BY 4.0 licence at DOI:10.5281/zenodo.4694518
Graph Names:
The RDF graphs have a common name-space: http://www.southgreen.fr/agrold/
The list of graphs are follows:
- http://www.southgreen.fr/agrold/qtaro.genes
- http://www.southgreen.fr/agrold/qtaro.qtl
- http://www.southgreen.fr/agrold/rap.msu
- http://www.southgreen.fr/agrold/greenphyldb
- http://www.southgreen.fr/agrold/uniprot.plants
- http://www.southgreen.fr/agrold/gramene.qtl
- http://www.southgreen.fr/agrold/tropgenedb
- http://www.southgreen.fr/agrold/ensembl.plants
- http://www.southgreen.fr/agrold/protein.annotations
- http://www.southgreen.fr/agrold/qtl.annotations
- http://www.southgreen.fr/agrold/msu.genes
- http://www.southgreen.fr/agrold/gramene.cyc
- http://www.southgreen.fr/agrold/sniplaydb
- http://www.southgreen.fr/agrold/oryzabase.genes
- http://www.southgreen.fr/agrold/ricenetdb
- http://www.southgreen.fr/agrold/rapdb
- http://www.southgreen.fr/agrold/genome.hub
- http://www.southgreen.fr/agrold/plantftdb
- http://www.southgreen.fr/agrold/interpro
- http://www.southgreen.fr/agrold/cegresources
- http://www.southgreen.fr/agrold/ncbitaxon
- http://www.southgreen.fr/agrold/faldo
- http://www.southgreen.fr/agrold/ino
- http://www.southgreen.fr/agrold/ro
- http://www.southgreen.fr/agrold/go
- http://www.southgreen.fr/agrold/vocabulary
- http://www.southgreen.fr/agrold/so
- http://www.southgreen.fr/agrold/sio
- http://www.southgreen.fr/agrold/eco
- http://www.southgreen.fr/agrold/pato
- http://www.southgreen.fr/agrold/po
- http://www.southgreen.fr/agrold/to
URIs:
To make AgroLD Linked Data compliant, de-referenceable stable URIs provided by Identifiers.org (http://identifiers.org/), Ontobee (www.ontobee.org) and canonical stable URIs provided UniprotKB are used.
Datasets that are not included in these registries, new URIs were minted the identifiers for these datasets take the form:
http://www.southgreen.fr/agrold/[resource_namespace]/[identifier]Example:
http://www.southgreen.fr/agrold/biocyc.pathway/CALVIN-PWY
Similarly, properties, would be of the form:
http://www.southgreen.fr/agrold/[vocabulary]/[property]Example:
http://www.southgreen.fr/agrold/vocabulary/expressed_in
